ClinLabGeneticist
ClinLabGeneticist is a tool to assist geneticist for diagnosis of pathogenic variants from WES data. More than 10 public known databases are used for variant annotation. In the new version 2.2, this tool works in windows environment (window 7, 8). The tool is free for academia and can be downloaded from ClinLabGeneticistVersion2.28. If you met some problem such as “‘Microsoft.ACE.OLEDB.12.0”, please install AccessDatabaseEngine. In the first running, suggest you open ‘wes.accdb’ and “Enable” the Access database, then close ‘wes.accdb’. You may need Guidline-of-ClinLabGeneticist.
For demo purpose, we provide two data sets: one real clinical data (1408019) and one simulation data (testing) . The real data can be downloaded here: 1408019-CompHet 1408019-DeNovo 1408019-Recessive 1408019-Secondary . 1408019 has been distributed and ready for annotation using a reviewer’s account. The “testing” data has reviewed by all reviewers and is ready for combining by administrator. We created 1 administrator and 5 reviewer accounts. Please email us for the passwords. [email protected]
1 Administrator account:
5 Reviewers accounts:
If you want to try everything from vcf file, please contact [email protected].
For input data, we suggest using file names with a hyphen to separate the case and variant type, for example, 1408019-DeNovo.txt. The text file is tab delimited. the variant data suggest to be filtered by Ingenuity Variants Analysis tool or other filtering tools based on the categories of the variants after called by GATK pipeline. If you don’t run through Ingenuity, make sure the data includes the following columns.
- Chromosome
- Position
- Reference Allele
- Sample Allele
- Gene Symbol
- Transcript ID
- Transcript Variant
- Protein Variant
- SIFT Function Prediction
- PolyPhen-2 Function Prediction
- Conservation phyloP p-value
- dbSNP ID
- 1000 Genomes Frequency
To accurately identify pathogenic variants, a hierarchical decision making strategy is made by ClinLabGeneticist. Variants called using GATK pipeline are import into Ingenuity Variants Analysis tool for further filtering and marked as recessive, de novo, dominant, compound het and secondary findings variants followed the guideline from ACMG. In the first level, reviewers who have solid clinical genetics background annotate the variants and assess their pathogenecity. Then, a group meeting based on the assessment from all reviewers discuss the discrepancy and make second level decision. Next, after validated by Sanger, directors meet to confirm and make final decision on each reportable variants. To achieve this aim, 2 roles are designed in ClinLabGeneticist. Administrators are responsible for assignment distribution, combination, and discussion. Reviewers start review assignments, make comments, and make decision on each variants assigned. Figure 1-6 are snapshots for the administrator dashboard. Figure 7-10 for the reviewer dashboard.

Figure 1. Administrator panel |
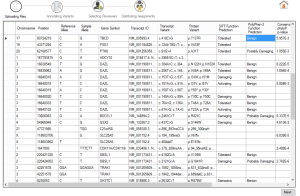
Figure 2. File uploading |
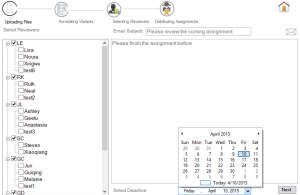
Figure 3. Reviewer selection |
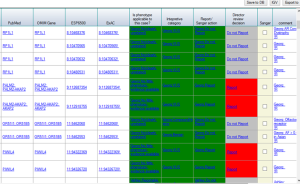
Figure 4. Assignments combination |
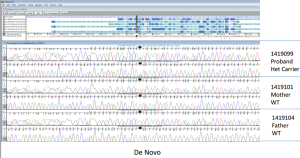
Figure 5. Variants validated by Sanger |
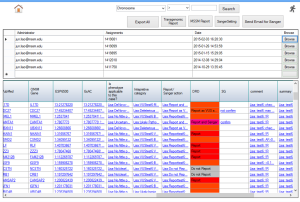
Figure 6. Reportable variants |

Figure 7. Reviewer dashboard |
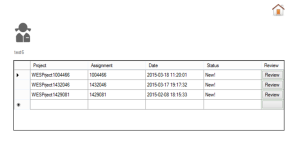
Figure 8. New assignments list |
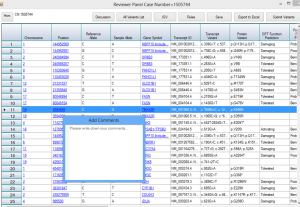
Figure 9. Master table to annotate and comment variants. |
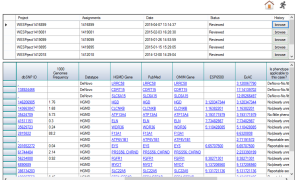
Figure 10. Browse the reviewed assignments |

